Structured State Space Models for Multiple Instance Learning in Digital Pathology
Leo Fillioux, Joseph Boyd, Maria Vakalopoulou, Paul-Henry Cournède, Stergios Christodoulidis
Early Accept, MICCAI 2023
[code]
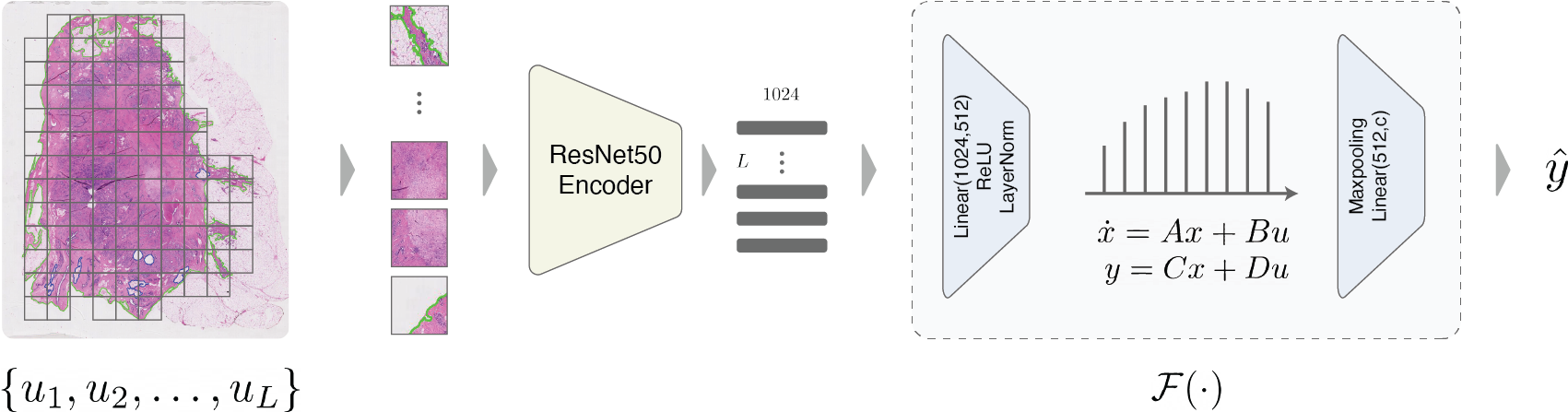
Abstract: Multiple instance learning is an ideal mode of analysis for histopathology data, where vast whole slide images are typically annotated with a single global label. In such cases, a whole slide image is modeled as a collection of tissue patches to be aggregated and classified. Common models for performing this classification include recurrent neural networks and transformers. Although powerful compression algorithms, such as deep pre-trained neural networks, are used to reduce the dimensionality of each patch, the sequences arising from whole slide images remain excessively long, routinely containing tens of thousands of patches. Structured state space models are an emerging alternative for sequence modelling, specifically designed for the efficient modelling of long sequences. These models invoke an optimal projection of an input sequence into memory units that compress the entire sequence. In this paper, we propose the use of state space models as a multiple instance learner to a variety of problems in digital pathology. Across experiments in metastasis detection, cancer subtyping, mutation classification, and multitask learning, we demonstrate the competitiveness of this new class of models with existing state of the art approaches.